I’ve spent so many years using and broadcasting my love for R and using Python quite minimally. Having read recently about machine learning in Python, I decided to take on a fun little ML project using Python from start to finish.
What follows below takes advantage of a neat dataset from the UCI Machine Learning Repository. The data contain Math test performance of 649 students in 2 Portuguese schools. What’s neat about this data set is that in addition to grades on the students’ 3 Math tests, they managed to collect a whole whack of demographic variables (and some behavioural) as well. That lead me to the question of how well can you predict final math test performance based on demographics and behaviour alone. In other words, who is likely to do well, and who is likely to tank?
I have to admit before I continue, I initially intended on doing this analysis in Python alone, but I actually felt lost 3 quarters of the way through and just did the whole darned thing in R. Once I had completed the analysis in R to my liking, I then went back to my Python analysis and continued until I finished to my reasonable satisfaction. For that reason, for each step in the analysis, I will show you the code I used in Python, the results, and then the same thing in R. Do not treat this as a comparison of Python’s machine learning capabilities versus R per se. Please treat this as a comparison of my understanding of how to do machine learning in Python versus R!
Without further ado, let’s start with some import statements in Python and library statements in R:
#Python Code
from pandas import *
from matplotlib import *
import seaborn as sns
sns.set_style("darkgrid")
import matplotlib.pyplot as plt
%matplotlib inline # I did this in ipython notebook, this makes the graphs show up inline in the notebook.
import statsmodels.formula.api as smf
from scipy import stats
from numpy.random import uniform
from numpy import arange
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error
from math import sqrt
mat_perf = read_csv('/home/inkhorn/Student Performance/student-mat.csv', delimiter=';')
I’d like to comment on the number of import statements I found myself writing in this python script. Eleven!! Is that even normal? Note the smaller number of library statements in my R code block below:
#R Code
library(ggplot2)
library(dplyr)
library(ggthemr)
library(caret)
ggthemr('flat') # I love ggthemr!
mat_perf = read.csv('student-mat.csv', sep = ';')
Now let’s do a quick plot of our target variable, scores on the students’ final math test, named ‘G3′.
#Python Code
sns.set_palette("deep", desat=.6)
sns.set_context(context='poster', font_scale=1)
sns.set_context(rc={"figure.figsize": (8, 4)})
plt.hist(mat_perf.G3)
plt.xticks(range(0,22,2))
Distribution of Final Math Test Scores (“G3″)
![Python Hist - G3]()
That looks pretty pleasing to my eyes. Now let’s see the code for the same thing in R (I know, the visual theme is different. So sue me!)
#R Code
ggplot(mat_perf) + geom_histogram(aes(x=G3), binwidth=2)
![Hist - G3]()
You’ll notice that I didn’t need to tweak any palette or font size parameters for the R plot, because I used the very fun ggthemr package. You choose the visual theme you want, declare it early on, and then all subsequent plots will share the same theme! There is a command I’ve hidden, however, modifying the figure height and width. I set the figure size using rmarkdown, otherwise I just would have sized it manually using the export menu in the plot frame in RStudio. I think both plots look pretty nice, although I’m very partial to working with ggthemr!
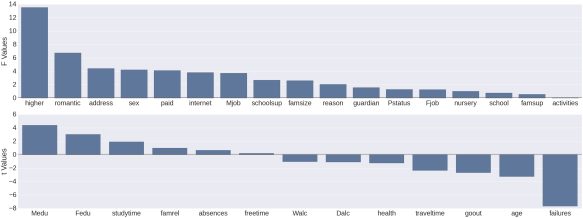
Univariate estimates of variable importance for feature selection
Below, what I’ve done in both languages is to cycle through each variable in the dataset (excepting prior test scores) insert the variable name in a dictionary/list, and get a measure of importance of how predictive that variable is, alone, of the final math test score (variable G3). Of course if the variable is qualitative then I get an F score from an ANOVA, and if it’s quantitative then I get a t score from the regression.
In the case of Python this is achieved in both cases using the ols function from scipy’s statsmodels package. In the case of R I’ve achieved this using the aov function for qualitative and the lm function for quantitative variables. The numerical outcome, as you’ll see from the graphs, is the same.
#Python Code
test_stats = {'variable': [], 'test_type' : [], 'test_value' : []}
for col in mat_perf.columns[:-3]:
test_stats['variable'].append(col)
if mat_perf[col].dtype == 'O':
# Do ANOVA
aov = smf.ols(formula='G3 ~ C(' + col + ')', data=mat_perf, missing='drop').fit()
test_stats['test_type'].append('F Test')
test_stats['test_value'].append(round(aov.fvalue,2))
else:
# Do correlation
print col + 'n'
model = smf.ols(formula='G3 ~ ' + col, data=mat_perf, missing='drop').fit()
value = round(model.tvalues[1],2)
test_stats['test_type'].append('t Test')
test_stats['test_value'].append(value)
test_stats = DataFrame(test_stats)
test_stats.sort(columns='test_value', ascending=False, inplace=True)
#R Code
test.stats = list(test.type = c(), test.value = c(), variable = c())
for (i in 1:30) {
test.stats$variable[i] = names(mat_perf)[i]
if (is.factor(mat_perf[,i])) {
anova = summary(aov(G3 ~ mat_perf[,i], data=mat_perf))
test.stats$test.type[i] = "F test"
test.stats$test.value[i] = unlist(anova)[7]
}
else {
reg = summary(lm(G3 ~ mat_perf[,i], data=mat_perf))
test.stats$test.type[i] = "t test"
test.stats$test.value[i] = reg$coefficients[2,3]
}
}
test.stats.df = arrange(data.frame(test.stats), desc(test.value))
test.stats.df$variable = reorder(test.stats.df$variable, -test.stats.df$test.value)
And now for the graphs. Again you’ll see a bit more code for the Python graph vs the R graph. Perhaps someone will be able to show me code that doesn’t involve as many lines, or maybe it’s just the way things go with graphing in Python. Feel free to educate me :)
#Python Code
f, (ax1, ax2) = plt.subplots(2,1, figsize=(48,18), sharex=False)
sns.set_context(context='poster', font_scale=1)
sns.barplot(x='variable', y='test_value', data=test_stats.query("test_type == 'F Test'"), hline=.1, ax=ax1, x_order=[x for x in test_stats.query("test_type == 'F Test'")['variable']])
ax1.set_ylabel('F Values')
ax1.set_xlabel('')
sns.barplot(x='variable', y='test_value', data=test_stats.query("test_type == 't Test'"), hline=.1, ax=ax2, x_order=[x for x in test_stats.query("test_type == 't Test'")['variable']])
ax2.set_ylabel('t Values')
ax2.set_xlabel('')
sns.despine(bottom=True)
plt.tight_layout(h_pad=3)
![Python Bar Plot - Univariate Estimates of Variable Importance]()
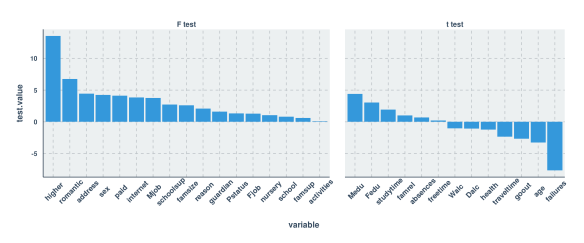
#R Code
ggplot(test.stats.df, aes(x=variable, y=test.value)) +
geom_bar(stat="identity") +
facet_grid(.~test.type , scales="free", space = "free") +
theme(axis.text.x = element_text(angle = 45, vjust=.75, size=11))
![Bar plot - Univariate Estimates of Variable Importance]()
As you can see, the estimates that I generated in both languages were thankfully the same. My next thought was to use only those variables with a test value (F or t) of 3.0 or higher. What you’ll see below is that this led to a pretty severe decrease in predictive power compared to being liberal with feature selection.
In reality, the feature selection I use below shouldn’t be necessary at all given the size of the data set vs the number of predictors, and the statistical method that I’m using to predict grades (random forest). What’s more is that my feature selection method in fact led me to reject certain variables which I later found to be important in my expanded models! For this reason it would be nice to investigate a scalable multivariate feature selection method (I’ve been reading a bit about boruta but am skeptical about how well it scales up) to have in my tool belt. Enough blathering, and on with the model training:
Training the First Random Forest Model
#Python code
usevars = [x for x in test_stats.query("test_value >= 3.0 | test_value <= -3.0")['variable']]
mat_perf['randu'] = np.array([uniform(0,1) for x in range(0,mat_perf.shape[0])])
mp_X = mat_perf[usevars]
mp_X_train = mp_X[mat_perf['randu'] <= .67]
mp_X_test = mp_X[mat_perf['randu'] > .67]
mp_Y_train = mat_perf.G3[mat_perf['randu'] <= .67]
mp_Y_test = mat_perf.G3[mat_perf['randu'] > .67]
# for the training set
cat_cols = [x for x in mp_X_train.columns if mp_X_train[x].dtype == "O"]
for col in cat_cols:
new_cols = get_dummies(mp_X_train[col])
new_cols.columns = col + '_' + new_cols.columns
mp_X_train = concat([mp_X_train, new_cols], axis=1)
# for the testing set
cat_cols = [x for x in mp_X_test.columns if mp_X_test[x].dtype == "O"]
for col in cat_cols:
new_cols = get_dummies(mp_X_test[col])
new_cols.columns = col + '_' + new_cols.columns
mp_X_test = concat([mp_X_test, new_cols], axis=1)
mp_X_train.drop(cat_cols, inplace=True, axis=1)
mp_X_test.drop(cat_cols, inplace=True, axis=1)
rf = RandomForestRegressor(bootstrap=True,
criterion='mse', max_depth=2, max_features='auto',
min_density=None, min_samples_leaf=1, min_samples_split=2,
n_estimators=100, n_jobs=1, oob_score=True, random_state=None,
verbose=0)
rf.fit(mp_X_train, mp_Y_train)
After I got past the part where I constructed the training and testing sets (with “unimportant” variables filtered out) I ran into a real annoyance. I learned that categorical variables need to be converted to dummy variables before you do the modeling (where each level of the categorical variable gets its own variable containing 1s and 0s. 1 means that the level was present in that row and 0 means that the level was not present in that row; so called “one-hot encoding”). I suppose you could argue that this puts less computational demand on the modeling procedures, but when you’re dealing with tree based ensembles I think this is a drawback. Let’s say you have a categorical variable with 5 features, “a” through “e”. It just so happens that when you compare a split on that categorical variable where “abc” is on one side and “de” is on the other side, there is a very significant difference in the dependent variable. How is one-hot encoding going to capture that? And then, your dataset which had a certain number of columns now has 5 additional columns due to the encoding. “Blah” I say!
Anyway, as you can see above, I used the get_dummies function in order to do the one-hot encoding. Also, you’ll see that I’ve assigned two thirds of the data to the training set and one third to the testing set. Now let’s see the same steps in R:
#R Code
keep.vars = match(filter(test.stats.df, abs(test.value) >= 3)$variable, names(mat_perf))
ctrl = trainControl(method="repeatedcv", number=10, selectionFunction = "oneSE")
mat_perf$randu = runif(395)
test = mat_perf[mat_perf$randu > .67,]
trf = train(mat_perf[mat_perf$randu <= .67,keep.vars], mat_perf$G3[mat_perf$randu <= .67],
method="rf", metric="RMSE", data=mat_perf,
trControl=ctrl, importance=TRUE)
Wait a minute. Did I really just train a Random Forest model in R, do cross validation, and prepare a testing data set with 5 commands!?!? That was a lot easier than doing these preparations and not doing cross validation in Python! I did in fact try to figure out cross validation in sklearn, but then I was having problems accessing variable importances after. I do like the caret package :) Next, let’s see how each of the models did on their testing set:
Testing the First Random Forest Model
#Python Code
y_pred = rf.predict(mp_X_test)
sns.set_context(context='poster', font_scale=1)
first_test = DataFrame({"pred.G3.keepvars" : y_pred, "G3" : mp_Y_test})
sns.lmplot("G3", "pred.G3.keepvars", first_test, size=7, aspect=1.5)
print 'r squared value of', stats.pearsonr(mp_Y_test, y_pred)[0]**2
print 'RMSE of', sqrt(mean_squared_error(mp_Y_test, y_pred))
![Python Scatter Plot - First Model Pred vs Actual]()
R^2 value of 0.104940038879
RMSE of 4.66552400292
Here, as in all cases when making a prediction using sklearn, I use the predict method to generate the predicted values from the model using the testing set and then plot the prediction (“pred.G3.keepvars”) vs the actual values (“G3″) using the lmplot function. I like the syntax that the lmplot function from the seaborn package uses as it is simple and familiar to me from the R world (where the arguments consist of “X variable, Y Variable, dataset name, other aesthetic arguments). As you can see from the graph above and from the R^2 value, this model kind of sucks. Another thing I like here is the quality of the graph that seaborn outputs. It’s nice! It looks pretty modern, the text is very readable, and nothing looks edgy or pixelated in the plot. Okay, now let’s look at the code and output in R, using the same predictors.
#R Code
test$pred.G3.keepvars = predict(trf, test, "raw")
cor.test(test$G3, test$pred.G3.keepvars)$estimate[[1]]^2
summary(lm(test$G3 ~ test$pred.G3.keepvars))$sigma
ggplot(test, aes(x=G3, y=pred.G3.keepvars)) + geom_point() + stat_smooth(method="lm") + scale_y_continuous(breaks=seq(0,20,4), limits=c(0,20))
![Scatter Plot - First Model Pred vs Actual]()
R^2 value of 0.198648
RMSE of 4.148194
Well, it looks like this model sucks a bit less than the Python one. Quality-wise, the plot looks super nice (thanks again, ggplot2 and ggthemr!) although by default the alpha parameter is not set to account for overplotting. The docs page for ggplot2 suggests setting alpha=.05, but for this particular data set, setting it to .5 seems to be better.
Finally for this section, let’s look at the variable importances generated for each training model:
#Python Code
importances = DataFrame({'cols':mp_X_train.columns, 'imps':rf.feature_importances_})
print importances.sort(['imps'], ascending=False)
cols imps
3 failures 0.641898
0 Medu 0.064586
10 sex_F 0.043548
19 Mjob_services 0.038347
11 sex_M 0.036798
16 Mjob_at_home 0.036609
2 age 0.032722
1 Fedu 0.029266
15 internet_yes 0.016545
6 romantic_no 0.013024
7 romantic_yes 0.011134
5 higher_yes 0.010598
14 internet_no 0.007603
4 higher_no 0.007431
12 paid_no 0.002508
20 Mjob_teacher 0.002476
13 paid_yes 0.002006
18 Mjob_other 0.001654
17 Mjob_health 0.000515
8 address_R 0.000403
9 address_U 0.000330
#R Code
varImp(trf)
## rf variable importance
##
## Overall
## failures 100.000
## romantic 49.247
## higher 27.066
## age 17.799
## Medu 14.941
## internet 12.655
## sex 8.012
## Fedu 7.536
## Mjob 5.883
## paid 1.563
## address 0.000
My first observation is that it was obviously easier for me to get the variable importances in R than it was in Python. Next, you’ll certainly see the symptom of the dummy coding I had to do for the categorical variables. That’s no fun, but we’ll survive through this example analysis, right? Now let’s look which variables made it to the top:
Whereas failures, mother’s education level, sex and mother’s job made it to the top of the list for the Python model, the top 4 were different apart from failures in the R model.
With the understanding that the variable selection method that I used was inappropriate, let’s move on to training a Random Forest model using all predictors except the prior 2 test scores. Since I’ve already commented above on my thoughts about the various steps in the process, I’ll comment only on the differences in results in the remaining sections.
Training and Testing the Second Random Forest Model
#Python Code
#aav = almost all variables
mp_X_aav = mat_perf[mat_perf.columns[0:30]]
mp_X_train_aav = mp_X_aav[mat_perf['randu'] <= .67]
mp_X_test_aav = mp_X_aav[mat_perf['randu'] > .67]
# for the training set
cat_cols = [x for x in mp_X_train_aav.columns if mp_X_train_aav[x].dtype == "O"]
for col in cat_cols:
new_cols = get_dummies(mp_X_train_aav[col])
new_cols.columns = col + '_' + new_cols.columns
mp_X_train_aav = concat([mp_X_train_aav, new_cols], axis=1)
# for the testing set
cat_cols = [x for x in mp_X_test_aav.columns if mp_X_test_aav[x].dtype == "O"]
for col in cat_cols:
new_cols = get_dummies(mp_X_test_aav[col])
new_cols.columns = col + '_' + new_cols.columns
mp_X_test_aav = concat([mp_X_test_aav, new_cols], axis=1)
mp_X_train_aav.drop(cat_cols, inplace=True, axis=1)
mp_X_test_aav.drop(cat_cols, inplace=True, axis=1)
rf_aav = RandomForestRegressor(bootstrap=True,
criterion='mse', max_depth=2, max_features='auto',
min_density=None, min_samples_leaf=1, min_samples_split=2,
n_estimators=100, n_jobs=1, oob_score=True, random_state=None,
verbose=0)
rf_aav.fit(mp_X_train_aav, mp_Y_train)
y_pred_aav = rf_aav.predict(mp_X_test_aav)
second_test = DataFrame({"pred.G3.almostallvars" : y_pred_aav, "G3" : mp_Y_test})
sns.lmplot("G3", "pred.G3.almostallvars", second_test, size=7, aspect=1.5)
print 'r squared value of', stats.pearsonr(mp_Y_test, y_pred_aav)[0]**2
print 'RMSE of', sqrt(mean_squared_error(mp_Y_test, y_pred_aav))
![Python Scatter Plot - Second Model Pred vs Actual]()
R^2 value of 0.226587731888
RMSE of 4.3338674965
Compared to the first Python model, the R^2 on this one is more than doubly higher (the first R^2 was .10494) and the RMSE is 7.1% lower (the first was 4.6666254). The predicted vs actual plot confirms that the predictions still don’t look fantastic compared to the actuals, which is probably the main reason why the RMSE hasn’t decreased by so much. Now to the R code using the same predictors:
#R code
trf2 = train(mat_perf[mat_perf$randu <= .67,1:30], mat_perf$G3[mat_perf$randu <= .67],
method="rf", metric="RMSE", data=mat_perf,
trControl=ctrl, importance=TRUE)
test$pred.g3.almostallvars = predict(trf2, test, "raw")
cor.test(test$G3, test$pred.g3.almostallvars)$estimate[[1]]^2
summary(lm(test$G3 ~ test$pred.g3.almostallvars))$sigma
ggplot(test, aes(x=G3, y=pred.g3.almostallvars)) + geom_point() +
stat_smooth() + scale_y_continuous(breaks=seq(0,20,4), limits=c(0,20))
![Scatter Plot - Second Model Pred vs Actual]()
R^2 value of 0.3262093
RMSE of 3.8037318
Compared to the first R model, the R^2 on this one is approximately 1.64 times higher (the first R^2 was .19865) and the RMSE is 8.3% lower (the first was 4.148194). Although this particular model is indeed doing better at predicting values in the test set than the one built in Python using the same variables, I would still hesitate to assume that the process is inherently better for this data set. Due to the randomness inherent in Random Forests, one run of the training could be lucky enough to give results like the above, whereas other times the results might even be slightly worse than what I managed to get in Python. I confirmed this, and in fact most additional runs of this model in R seemed to result in an R^2 of around .20 and an RMSE of around 4.2.
Again, let’s look at the variable importances generated for each training model:
#Python Code
importances_aav = DataFrame({'cols':mp_X_train_aav.columns, 'imps':rf_aav.feature_importances_})
print importances_aav.sort(['imps'], ascending=False)
cols imps
5 failures 0.629985
12 absences 0.057430
1 Medu 0.037081
41 schoolsup_yes 0.036830
0 age 0.029672
23 Mjob_at_home 0.029642
16 sex_M 0.026949
15 sex_F 0.026052
40 schoolsup_no 0.019097
26 Mjob_services 0.016354
55 romantic_yes 0.014043
51 higher_yes 0.012367
2 Fedu 0.011016
39 guardian_other 0.010715
37 guardian_father 0.006785
8 goout 0.006040
11 health 0.005051
54 romantic_no 0.004113
7 freetime 0.003702
3 traveltime 0.003341
#R Code
varImp(trf2)
## rf variable importance
##
## only 20 most important variables shown (out of 30)
##
## Overall
## absences 100.00
## failures 70.49
## schoolsup 47.01
## romantic 32.20
## Pstatus 27.39
## goout 26.32
## higher 25.76
## reason 24.02
## guardian 22.32
## address 21.88
## Fedu 20.38
## school 20.07
## traveltime 20.02
## studytime 18.73
## health 18.21
## Mjob 17.29
## paid 15.67
## Dalc 14.93
## activities 13.67
## freetime 12.11
Now in both cases we’re seeing that absences and failures are considered as the top 2 most important variables for predicting final math exam grade. It makes sense to me, but frankly is a little sad that the two most important variables are so negative :( On to to the third Random Forest model, containing everything from the second with the addition of the students’ marks on their second math exam!
Training and Testing the Third Random Forest Model
#Python Code
#allv = all variables (except G1)
allvars = range(0,30)
allvars.append(31)
mp_X_allv = mat_perf[mat_perf.columns[allvars]]
mp_X_train_allv = mp_X_allv[mat_perf['randu'] <= .67]
mp_X_test_allv = mp_X_allv[mat_perf['randu'] > .67]
# for the training set
cat_cols = [x for x in mp_X_train_allv.columns if mp_X_train_allv[x].dtype == "O"]
for col in cat_cols:
new_cols = get_dummies(mp_X_train_allv[col])
new_cols.columns = col + '_' + new_cols.columns
mp_X_train_allv = concat([mp_X_train_allv, new_cols], axis=1)
# for the testing set
cat_cols = [x for x in mp_X_test_allv.columns if mp_X_test_allv[x].dtype == "O"]
for col in cat_cols:
new_cols = get_dummies(mp_X_test_allv[col])
new_cols.columns = col + '_' + new_cols.columns
mp_X_test_allv = concat([mp_X_test_allv, new_cols], axis=1)
mp_X_train_allv.drop(cat_cols, inplace=True, axis=1)
mp_X_test_allv.drop(cat_cols, inplace=True, axis=1)
rf_allv = RandomForestRegressor(bootstrap=True,
criterion='mse', max_depth=2, max_features='auto',
min_density=None, min_samples_leaf=1, min_samples_split=2,
n_estimators=100, n_jobs=1, oob_score=True, random_state=None,
verbose=0)
rf_allv.fit(mp_X_train_allv, mp_Y_train)
y_pred_allv = rf_allv.predict(mp_X_test_allv)
third_test = DataFrame({"pred.G3.plusG2" : y_pred_allv, "G3" : mp_Y_test})
sns.lmplot("G3", "pred.G3.plusG2", third_test, size=7, aspect=1.5)
print 'r squared value of', stats.pearsonr(mp_Y_test, y_pred_allv)[0]**2
print 'RMSE of', sqrt(mean_squared_error(mp_Y_test, y_pred_allv))
![Python Scatter Plot - Third Model Pred vs Actual]()
R^2 value of 0.836089929903
RMSE of 2.11895794845
Obviously we have added a highly predictive piece of information here by adding the grades from their second math exam (variable name was “G2″). I was reluctant to add this variable at first because when you predict test marks with previous test marks then it prevents the model from being useful much earlier on in the year when these tests have not been administered. However, I did want to see what the model would look like when I included it anyway! Now let’s see how predictive these variables were when I put them into a model in R:
#R Code
trf3 = train(mat_perf[mat_perf$randu <= .67,c(1:30,32)], mat_perf$G3[mat_perf$randu <= .67],
method="rf", metric="RMSE", data=mat_perf,
trControl=ctrl, importance=TRUE)
test$pred.g3.plusG2 = predict(trf3, test, "raw")
cor.test(test$G3, test$pred.g3.plusG2)$estimate[[1]]^2
summary(lm(test$G3 ~ test$pred.g3.plusG2))$sigma
ggplot(test, aes(x=G3, y=pred.g3.plusG2)) + geom_point() +
stat_smooth(method="lm") + scale_y_continuous(breaks=seq(0,20,4), limits=c(0,20))
![Scatter Plot - Third Model Pred vs Actual]()
R^2 value of 0.9170506
RMSE of 1.3346087
Well, it appears that yet again we have a case where the R model has fared better than the Python model. I find it notable that when you look at the scatterplot for the Python model you can see what look like steps in the points as you scan your eyes from the bottom-left part of the trend line to the top-right part. It appears that the Random Forest model in R has benefitted from the tuning process and as a result the distribution of the residuals are more homoscedastic and also obviously closer to the regression line than the Python model. I still wonder how much more similar these results would be if I had carried out the Python analysis by tuning while cross validating like I did in R!
For the last time, let’s look at the variable importances generated for each training model:
#Python Code
importances_allv = DataFrame({'cols':mp_X_train_allv.columns, 'imps':rf_allv.feature_importances_})
print importances_allv.sort(['imps'], ascending=False)
cols imps
13 G2 0.924166
12 absences 0.075834
14 school_GP 0.000000
25 Mjob_health 0.000000
24 Mjob_at_home 0.000000
23 Pstatus_T 0.000000
22 Pstatus_A 0.000000
21 famsize_LE3 0.000000
20 famsize_GT3 0.000000
19 address_U 0.000000
18 address_R 0.000000
17 sex_M 0.000000
16 sex_F 0.000000
15 school_MS 0.000000
56 romantic_yes 0.000000
27 Mjob_services 0.000000
11 health 0.000000
10 Walc 0.000000
9 Dalc 0.000000
8 goout 0.000000
#R Code
varImp(trf3)
## rf variable importance
##
## only 20 most important variables shown (out of 31)
##
## Overall
## G2 100.000
## absences 33.092
## failures 9.702
## age 8.467
## paid 7.591
## schoolsup 7.385
## Pstatus 6.604
## studytime 5.963
## famrel 5.719
## reason 5.630
## guardian 5.278
## Mjob 5.163
## school 4.905
## activities 4.532
## romantic 4.336
## famsup 4.335
## traveltime 4.173
## Medu 3.540
## Walc 3.278
## higher 3.246
Now this is VERY telling, and gives me insight as to why the scatterplot from the Python model had that staircase quality to it. The R model is taking into account way more variables than the Python model. G2 obviously takes the cake in both models, but I suppose it overshadowed everything else by so much in the Python model, that for some reason it just didn’t find any use for any other variable than absences.
Conclusion
This was fun! For all the work I did in Python, I used IPython Notebook. Being an avid RStudio user, I’m not used to web-browser based interactive coding like what IPython Notebook provides. I discovered that I enjoy it and found it useful for laying out the information that I was using to write this blog post (I also laid out the R part of this analysis in RMarkdown for that same reason). What I did not like about IPython Notebook is that when you close it/shut it down/then later reinitialize it, all of the objects that form your data and analysis are gone and all you have left are the results. You must then re-run all of your code so that your objects are resident in memory again. It would be nice to have some kind of convenience function to save everything to disk so that you can reload at a later time.
I found myself stumbling a lot trying to figure out which Python packages to use for each particular purpose and I tended to get easily frustrated. I had to keep reminding myself that it’s a learning curve to a similar extent as it was for me while I was learning R. This frustration should not be a deterrent from picking it up and learning how to do machine learning in Python. Another part of my frustration was not being able to get variable importances from my Random Forest models in Python when I was building them using cross validation and grid searches. If you have a link to share with me that shows an example of this, I’d be happy to read it.
I liked seaborn and I think if I spend more time with it then perhaps it could serve as a decent alternative to graphing in ggplot2. That being said, I’ve spent so much time using ggplot2 that sometimes I wonder if there is anything out there that rivals its flexibility and elegance!
The issue I mentioned above with categorical variables is annoying and it really makes me wonder if using a Tree based R model would intrinsically be superior due to its automatic handling of categorical variables compared with Python, where you need to one-hot encode these variables.
All in all, I hope this was as useful and educational for you as it was for me. It’s important to step outside of your comfort zone every once in a while :)
![]()
![]()
To
leave a comment for the author, please follow the link and comment on his blog:
Data Until I Die!.
R-bloggers.com offers
daily e-mail updates about
R news and
tutorials on topics such as: visualization (
ggplot2,
Boxplots,
maps,
animation), programming (
RStudio,
Sweave,
LaTeX,
SQL,
Eclipse,
git,
hadoop,
Web Scraping) statistics (
regression,
PCA,
time series,
trading) and more...